Clarion2: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 51: | Line 51: | ||
[[File:GEAN4 simulation of 152Eu.png|600px|thumb|none|GEANT4 simulation of 152Eu]] | [[File:GEAN4 simulation of 152Eu.png|600px|thumb|none|GEANT4 simulation of 152Eu]] | ||
A GEANT4 simulation for Clarion2 can be found at https://fsunuc.physics.fsu.edu/git/pderosa888/CLARION2_GEANT4. | A GEANT4 simulation for Clarion2 can be found at https://fsunuc.physics.fsu.edu/git/pderosa888/CLARION2_GEANT4. Please ask Raeed Nawaf for an updated model here: rn24b@fsu.edu | ||
[[File:Clarion2 model.png|600px|thumb|none|Clarion2 GEANT4 model]] | <!-- [[File:Clarion2 model.png|600px|thumb|none|Clarion2 GEANT4 model]] --> | ||
[[File:Clarion02 Geant With BGO.png|600px|thumb|none|Clarion02 Geant With BGO]] | |||
[[File:Clarion02 Geant With BGO.png|600px|thumb|Clarion02 Geant With BGO]] | |||
== Trinity == | == Trinity == | ||
| Line 83: | Line 82: | ||
{{Notice | Detail of operation is needed.}} | {{Notice | Detail of operation is needed.}} | ||
==Casual Tutorial For Getting a Pixie Crate Up and Running== | |||
This is meant to be a bit of a tutorial on how to get a Pixie16 crate running from a fresh flash of Ubuntu 18. | |||
===Prerequisites=== | |||
Make sure that you have at an XIA crate with at least one Rev. F Pixie16 board. Rev. F boards can be recognized by the female headers on the face instead of the male. Rev. D boards have the male, there are several floating around Fox Lab. | |||
Make sure that you have a matching pair of optical communication boards: one to go in the crate and one to go into the PC. These model numbers should match or be close. For example, I had to use a PCI-8331/8336 board in the PC and a PXI-8336 board in the crate. It will not work if there is a mismatch. Trust me. | |||
Make sure you are running Ubuntu 18 with kernel 4.15. The drivers will work with this setup only. | |||
===Installation=== | |||
The install here is a bit of a lie. In your home directory copy the entire ~/pxi/ folder from the Clarion02 DAQ PC. All following instructions are on the local PC. | |||
Navigate to <code> cd ~/pxi/PlxSdk/ </code> | |||
Once here give the commands: | |||
<code> make clean </code> | |||
<code> make </code> | |||
This will build a bunch of stuff. | |||
Next, navigate to the Drivers folder: <code> cd ~/pxi/PlxSdk/Drivers </code> | |||
I had the most luck with the "builddrivers.sh" script. | |||
Start with the PCI drivers: <code> ./builddrivers Svc </code> | |||
This will likely throw some errors. If it does find the Dispatch.c script is it complaining about, probably here: ~/pxi/PlxSdk/Driver/PlxSvc/ | |||
Edit the includes to look like this: | |||
... I need to find the script so that I can copy the header.... Bad me | |||
= LeCroy HV Mainframe= | = LeCroy HV Mainframe= | ||
Latest revision as of 17:03, 3 February 2026
Introduction
Clarion2 [1] is the successor of Clarion [2] (CLover Array for Radioactive ION beam). It is a 4π gamma ray detectors array with 16 High-Purity Germanium (HPGe) clover detectors. Each HPGe detector is shied with a BGO (Bismuth Germanate [Bi4Ge3O12]) shield.
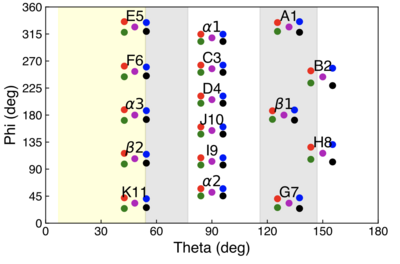
Each clover detector has 4 crystals, color-coded as Red, Blue, Green, and Black. Here is the angular map for all 16 clovers. Further details of the array can be found here or here.
| Solid work model needed |
HPGe Clover Detector
The detectors are produced by EURISYS Measure, now CANBERRA, owned by MIRION Technologies [1]. An approximate 3.3 kg clover detector consists of 4 coaxial crystals (dimension 50 mm in diameter, 80 mm in length) [Mirion webpage].
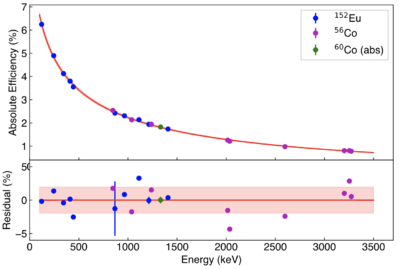
Typical photon-peak efficiency curve
The absolute photo-peak efficiency at 1332 keV is ~1.8%.
The equation for the absolute efficiency curve is
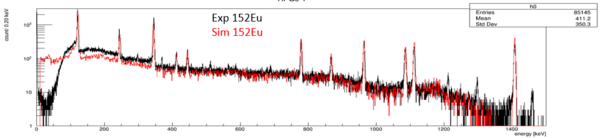
GEANT4 simulation
The performance of a single clover detector was simulated using GEANT4. The source code can be downloaded from https://github.com/goluckyryan/HPGeClover.
A GEANT4 simulation for Clarion2 can be found at https://fsunuc.physics.fsu.edu/git/pderosa888/CLARION2_GEANT4. Please ask Raeed Nawaf for an updated model here: rn24b@fsu.edu
Trinity
Trinity is formed by GAGG (Gadolinium Aluminium Gallium Garnet) scintillator crystals. It is located in the target chamber of the Clarion2. The GAGG crystals are arranged in 5 rings, covering 7-54 degrees. Currently only two rings are implemented, which cover 14-24 and 34-44 degrees respectively.
Neutron Shield
In 2022 April, a set of neutron shields surrounded Clarion2 for shielding slow neutrons.
Clarion2 private network
The Clarion2 uses its own private network. The network structure is illustrated as following:
Fox's Lab network --- clarion2(Mac, gateway) ---- 10 Gb switch
+--------- clarion2 DAQ
+--------- LN2 control, HV control
DAQ
The DAQ system is fully digitized with Pixie16 digitizer. The signals from the Clover and BGO are directly fed to the Pixie16 digitizers. The data processing library developed by Tim Gray can be found at https://github.com/belmakier/libpixie and https://github.com/belmakier/libClarionTrinity .
The Plx driver was modified from the Broadcom driver. The PixieSDK (legacy) was also modified.
| Detail of operation is needed. |
Casual Tutorial For Getting a Pixie Crate Up and Running
This is meant to be a bit of a tutorial on how to get a Pixie16 crate running from a fresh flash of Ubuntu 18.
Prerequisites
Make sure that you have at an XIA crate with at least one Rev. F Pixie16 board. Rev. F boards can be recognized by the female headers on the face instead of the male. Rev. D boards have the male, there are several floating around Fox Lab.
Make sure that you have a matching pair of optical communication boards: one to go in the crate and one to go into the PC. These model numbers should match or be close. For example, I had to use a PCI-8331/8336 board in the PC and a PXI-8336 board in the crate. It will not work if there is a mismatch. Trust me.
Make sure you are running Ubuntu 18 with kernel 4.15. The drivers will work with this setup only.
Installation
The install here is a bit of a lie. In your home directory copy the entire ~/pxi/ folder from the Clarion02 DAQ PC. All following instructions are on the local PC.
Navigate to cd ~/pxi/PlxSdk/
Once here give the commands:
make clean
make
This will build a bunch of stuff.
Next, navigate to the Drivers folder: cd ~/pxi/PlxSdk/Drivers
I had the most luck with the "builddrivers.sh" script.
Start with the PCI drivers: ./builddrivers Svc
This will likely throw some errors. If it does find the Dispatch.c script is it complaining about, probably here: ~/pxi/PlxSdk/Driver/PlxSvc/
Edit the includes to look like this:
... I need to find the script so that I can copy the header.... Bad me
LeCroy HV Mainframe
The clovers and BGOs bias is supplied by two LeCroy 1458 HV Mainframes. These are controlled via a serial connection to the LN02 PC.
Clearing Panic Status
A panic status seems to happen with the loss of power. If the system is in a Panic state, the red LEDs on both sides of the "PANIC OFF" button will be on. To clear the Panic status:
- Turn key to "Local"
- Press the "HV Off" button. This should clear the status and make the red panic lights go out.
- TURN THE KEY BACK TO REMOTE!!! This is important so we can control the system via the serial connection.
You will need to do this procedure for each mainframe.
LN2 cooling system
The liquid Nitrogen (LN2) filling system developed by Mitch Allmond (Oak Ridge) is hosted on the LN2-control computer under the account "ln@ln02".
Changing Alert List
To change the alert list, edit ~/scripts/run-send-alarms.sh with the relevant emails.
ln@ln02 Directory
Several important directories for using the LN2 system are listed below.
/gln
/gln is the most important directory, it is where the control program is located and for most problems is where you will need to go
gln_ui
gln_ui is the user interface to issue commands to the fill program
fill <det-list>- fill detectors <det-list> now
fill <det-list> after <number-of-minutes>(or <hours:minutes>)- wait to fill detectors <det-list> by time <hours:minutes>
cool <det-list>- cool down detectors
ton <det-list>- turn on (enable fill for) detectors
tof <det-list>- turn off (disable fill for) detectors
con det#[.temp#]- turn on checking of detector temps
cof det#[.temp#]- turn off checking of detector temps
abort- abort and disable all fills
load [fig-file-name]- load configuration from fig-file-name (default = gln.fig)
exit-master- exit gln_master process
clr- clear all errors for entire system
| Proper usage is to clear individual detectors |
clrd <det-list>- clear errors for specific detectors
clrs- clear all slot errors
clrm- clear all manifold errors
d- display status
dt- display times of all detectors & manifolds (formatted dates and times)
dts- display times of all detectors & manifolds (in seconds)
dc- display configuration (valve and ADC IDs, etc)
mon- enter monitor mode; type control-C to exit
quit- exit gln_ui process
hvln- useless command for backward compatibility
clear- clear screen
?orhelp- help text for commands
Low Level Commands
radc <#>- read ADC number <#>
opv <#>clv <#>- open and close valves
http://clarion02.physics.fsu.edu/~clarion02/index.html
List of Collaborators
The list may not be completed.
| Name | Affiliation |
|---|---|
| Mitch Allmond | ORNL |
| Tim Gray | ORNL |
| Toby King | ORNL |
| Ingo Wiedenhoever | FSU |
| Sam Tabor | FSU |
| Vandana Tripathi | FSU |
| Lagy Baby | FSU |
| Soumik Bhattacharya | FSU |
| Caleb Benetti | FSU |
| Catur Wibisono | FSU |
| Samuel Ajayi | FSU |
| Ryan Tang | FSU |
| Ram Yadav | South Carolina State University |
| James Christie | UTK |
Clover Inventory
| Clover Position / Name | "Owner" | Most Recent Pump | Heat Applied at Pump? |
|---|---|---|---|
| A1 | ORNL | ? | ? |
| B2 | ORNL | ? | ? |
| C3 | ORNL | ? | ? |
| D4 | ORNL | ? | ? |
| E5 | FSU | January 13, 2026 | ? |
| F6 | Not Real | ? | ? |
| G7 | FSU | January 13, 2026 | ? |
| H8 | ORNL | ? | ? |
| I9 | FSU | ? | ? |
| J10 | FSU | ? | ? |
| K11 | ORNL - Gone but not forgotten | ? | ? |
Contact
- Mitch Allmond mailto:allmondjm@ornl.gov
- Tim Gray mailto:graytj@ornl.gov
- Vandan Tripathi mailto:vtripath@fsu.edu
References
- ↑ T. J. Gray et. al, NIM A 1041, 167392 (2022) https://doi.org/10.1016/j.nima.2022.167392
- ↑ C. J. Gross et. al, NIM A 450, 12 (2000) https://doi.org/10.1016/S0168-9002(00)00159-5
![{\displaystyle \epsilon (e_{\gamma }{\text{[keV]}})=\epsilon _{0}\exp ^{\left(\left(A+Bx+Cx^{2}\right)^{-G}+\left(D+Ey+Fy^{2}\right)^{-G}\right)^{-1/G}},}](https://wikimedia.org/api/rest_v1/media/math/render/svg/888ee00bc37483ea67f94cc7c6ab1059d600aa1a)